A new analysis method to identify interacting proteins in the development of drugs
An analysis method for protein–protein interactions using proximity-dependent biotinylation for next-generation drugs
Protein degraders induce degradation of "neo-substrates". Protein degraders are considered to be the "next-generation drug" because of their ability to thoroughly degrade target proteins. These compounds hold great promise because they can target proteins that have been difficult to target so far. Therefore, understanding the proteins which interact with protein degraders is important for their development. In this study, we developed an analysis method to comprehensively detect protein degrader-dependent interactions.
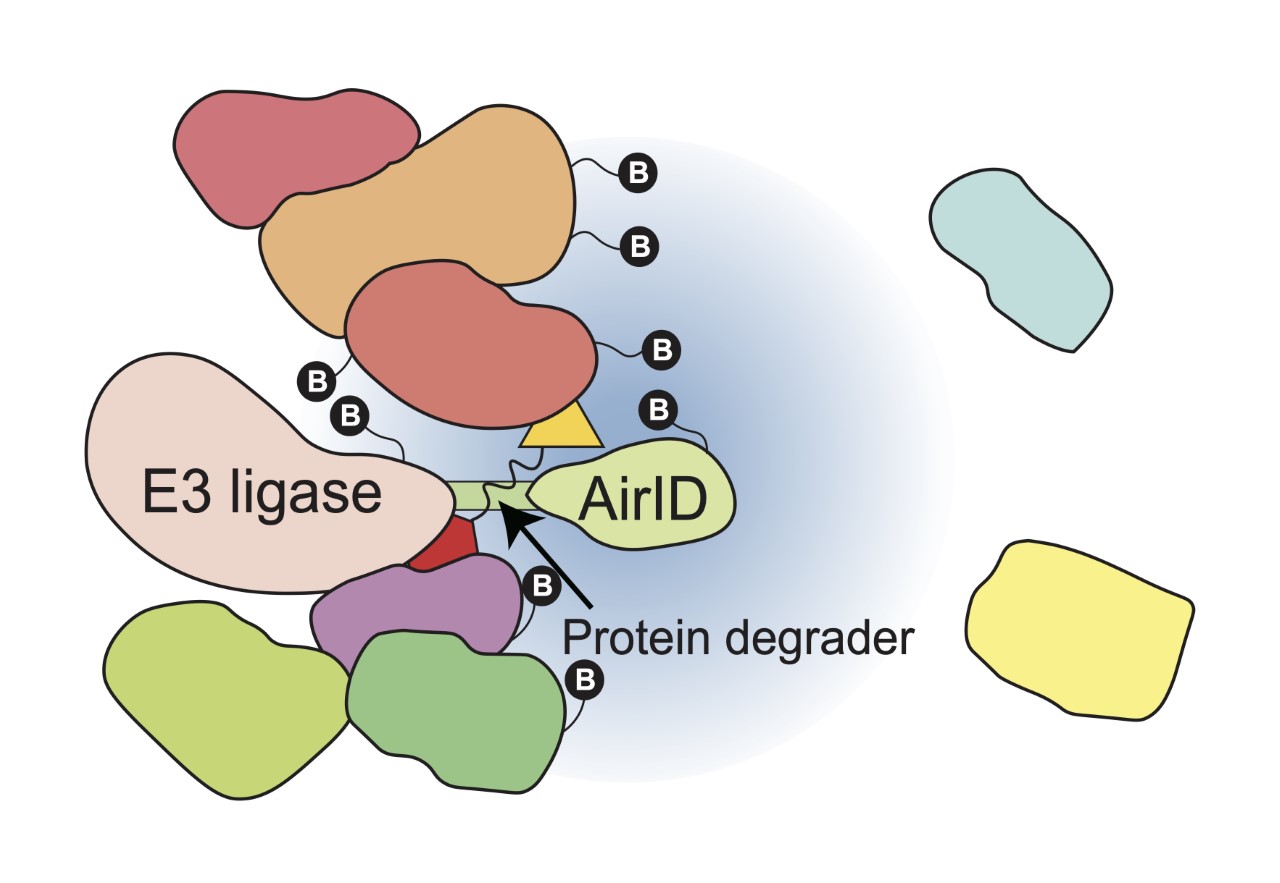
Protein degraders, which induce the protein degradation of target proteins, are expected to be the “next-generation drugs” because they can remove disease-related proteins from cells. Protein degraders, such as thalidomide and its derivatives (Immunomodulatory drugs/IMiDs), induce the degradation of specific proteins by binding to cereblon (CRBN), which is a component of the E3 ubiquitin ligase complex, a proteolytic enzyme. IMiDs function as a “molecular glue”, inducing protein degradation by recruiting target proteins to E3 ubiquitin ligase and are called molecular glue-type proteolytic inducers. Due to the clinical success of IMiDs, molecular glue-type protein degraders have become attractive compounds for use in combating many diseases. In addition, proteolysis targeting chimeras (PROTACs), which are chimeric compounds consisting of a compound that binds to an E3 ubiquitin ligase (E3 binder), such as IMiDs, and a compound that binds to a target protein (target binder), are now being developed. The development of PROTACs is expected to enable the targeting of proteins that have been difficult to target by therapeutic agents in the past. Therefore, the analysis of proteins that interact with E3 ubiquitin ligase and are induced to degrade is important for the development and clinical application of protein degraders. In 2020, this research group developed AirID (ancestral BirA for proximity-dependent biotin identification) as a proximal-dependent biotinylation labeling enzyme useful for protein–protein interaction analysis. Therefore, we aimed to develop a comprehensive analysis method to identify interacting proteins in a protein degrader-dependent manner.
In this study, we showed that AirID-fused E3 ubiquitin ligase, such as AirID-CRBN, can biotinylate target proteins of protein degraders in cells (Fig. 1). Furthermore, we showed that the enrichment of biotinylated peptides using the avidin-like protein tamabidine 2-REV is useful for the analysis of protein degrader-dependent interactions by mass spectrometry. Using this analysis method, we identified ZMYM2 (zinc finger MYM-type protein 2) as a target protein of pomalidomide, which is a thalidomide derivative. In addition, the ZMYM2-FGFR1 fusion protein that causes hematological cancer is also degraded by pomalidomide. Importantly, we confirmed that this analysis method can be used for known protein degraders such as Indisulam and PROTACs. These results indicate that the fusion of AirID with E3 ubiquitin ligase enables the comprehensive analysis of interacting proteins of protein degraders.
This analysis method using AirID can be used for the analysis of various protein degraders that will be developed in the future. Currently, various protein degraders are being actively developed for various diseases, and some of them are currently in the clinical stage. However, considering the potent effects of protein degraders, it is necessary to evaluate their interactions with proteins that are not intended to cause side effects. In fact, the results of this study indicated that various proteins interact with E3 ligase in a protein degrader-dependent manner. Based on these results, it is expected that the use of this analysis method will lead to the elucidation of the mechanism of action of protein degraders and the development of compounds that avoid side effects.
Reference URL: https://www.nature.com/articles/s41467-021-27818-z
Bibliographic Information
A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues, Satoshi Yamanaka, Yuto Horiuchi, Saya Matsuoka, Kohki Kido, Kohei Nishino, Mayaka Maeno, Norio Shibata, Hidetaka Kosako and Tatsuya Sawasaki, Nature Communications, 13, 183, doi: 10.1038/s41467-021-27818-z, 2022 (January 10).
Fundings
- This work was mainly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from the Japan Agency for Medical Research and Development (AMED) under Grant Number JP21am0101077 (T.S.)
- Project for Cancer Research and Therapeutic Evolution (P-CREATE) grant number (JP21cm0106181h0006 for S.Y.) from AMED
- Grant-in-Aid for Scientific Research on Innovative Areas (21H00285 for S.Y., JP16H06579 for T.S. and JP19H04966 for H.K) from the Japan Society for the Promotion of Science (JSPS). This work was also partially supported by JSPS KAKENHI (JP21K15076 for S.Y., JP19H03218 for T.S., and JP17H06112 for N.S.)
- Takeda Science Foundation
Media
Contact Person
Name : Satoshi Yamanaka
Phone : +81-89-927-8567
E-mail : yamanaka.satoshi.ze@ehime-u.ac.jp
Affiliation : Proteo-Science Center (PROS)