Discovering the Essential Role of SLFN11 in Cancer Treatment
Toward Future Applications in Precision Medicine
SLFN11 enhances the efficacy of DNA-damaging anticancer drugs by suppressing rRNA synthesis in the nucleolus, leading to rapid loss of the survival factor MCL1 and induction of apoptosis, even in TP53-deficient cells. This study uncovers a previously unknown mechanism by which SLFN11 drives cancer cell death and highlights its strong potential as a widely applicable biomarker for precision chemotherapy across multiple cancer types.
Cancer chemotherapy is widely used but often shows substantial variability in efficacy and causes significant adverse effects. Although predictive biomarkers are essential for optimizing treatment, most chemotherapeutic agents lack established markers. In 2012, the gene SLFN11 emerged as a strong predictor of sensitivity to DNA-damaging drugs, and clinical studies have shown that high SLFN11 expression correlates with better treatment response and survival across multiple cancers. However, its underlying mechanisms remained incompletely understood.
In this study, we discovered that SLFN11 strongly suppresses ribosomal RNA (rRNA) synthesis in the nucleolus upon exposure to DNA-damaging agents, leading to a rapid reduction in global protein synthesis. SLFN11-high cells exhibited a marked decline in RNA synthesis within hours of drug treatment and a dramatic loss of short-lived survival proteins such as MCL1, resulting in efficient induction of apoptosis. In contrast, SLFN11-deficient cells maintained rRNA synthesis and translation and failed to undergo apoptosis.
Importantly, this SLFN11-dependent cell-death pathway does not require p53, which is frequently inactivated in cancer. This mechanism represents a third, distinct mode of SLFN11-mediated cytotoxicity, complementing its previously reported roles in replication blockade (2018) and JNK pathway activation (2024), highlighting SLFN11 as a multifaceted driver of cancer cell death.
SLFN11 is highly expressed in approximately half of all cancers and can be readily assessed by routine immunohistochemistry, making it a practical and inexpensive biomarker. Unlike costly, one-time genomic testing, SLFN11 can be evaluated repeatedly and is applicable to the DNA-damaging chemotherapies used for a large proportion of patients. Future analysis using circulating tumor cells may enable even more dynamic and personalized treatment strategies.
Overall, this work provides major mechanistic insights into how SLFN11 enhances the efficacy of anticancer drugs and represents a significant step toward SLFN11-guided precision chemotherapy. Harnessing the full potential of SLFN11 may allow more patients to receive truly optimized cancer treatment.
Bibliographic Information
SLFN11-mediated ribosome biogenesis impairment induces TP53-independent apoptosis.
Akane Ogawa, Keiichi Izumikawa, Sota Tate, Sho Isoyama, Masaru Mori, Kohei Fujiwara, Soyoka Watanabe, Takayuki Ohga, Ukhyun Jo, Daiki Taniyama, Shojiro Kitajima, Soichiro Tanaka, Hiroshi Onji, Shun-Ichiro Kageyama, Gaku Yamamoto, Hitoshi Saito, Tomoko Yamamori Morita, Masayasu Okada, Manabu Natsumeda, Masami Nagahama, Junya Kobayashi, Akihiro Ohashi, Hiroyuki Sasanuma, Shigeki Higashiyama, Shingo Dan, Yves Pommier, Junko Murai#
2025・Molecular Cell・85(5)・894-912
DOI: 10.1016/j.molcel.2025.01.008
Fundings
- JST FOREST Program (Exploratory Research for Advanced Technology) (Grant No. JPMJFR2056, Junko Murai)
- JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (Grant Nos. JP19H03505 and JP23H02768, Junko Murai)
- JSPS KAKENHI Grant-in-Aid for Challenging Research (Exploratory) (Grant No. JP21K19415, Junko Murai)
Media
-
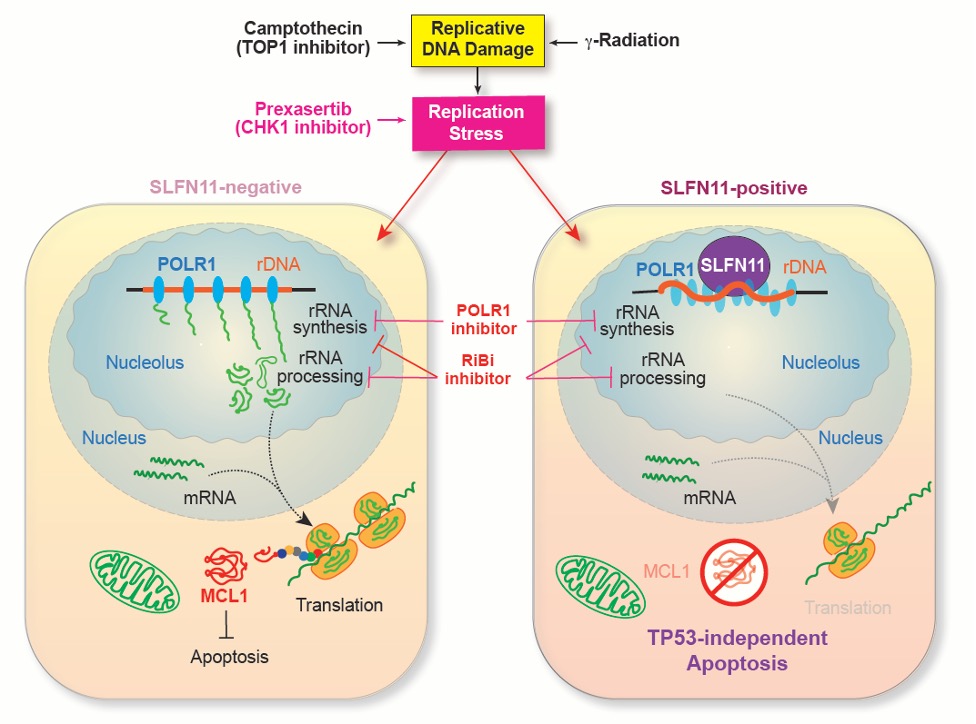
A Novel Apoptosis-Inducing Model Mediated by SLFN11 Through rRNA Synthesis Suppression and MCL1 Downregulation
Cells exposed to DNA damage, such as camptothecin or ionizing radiation, respond very differently depending on the presence of SLFN11.
In SLFN11-negative cells, rRNA synthesis and processing in the nucleolus are maintained, allowing translation to continue and preserving the survival factor MCL1, thereby suppressing apoptosis. In contrast, SLFN11-positive cells activate SLFN11 in response to DNA damage, leading to strong inhibition of rRNA synthesis by POLR1 (RNA polymerase I) within the nucleolus. This results in a global reduction in translation, rapid depletion of the short-lived protein MCL1, and subsequent induction of apoptosis.
This figure illustrates a novel mechanism by which SLFN11 enhances the cytotoxic effects of DNA-damaging anticancer agents.credit : Junko Murai
Usage Restriction : Please get copyright permission
Contact Person
Name : Junko Murai
Phone : +81 89-960-5254
E-mail : murai.junko.nk@ehime-u.ac.jp
Affiliation : Department of Cancer Control, Proteo-Science Center, Ehime University


