Innovative technology for the comprehensive analysis of membrane protein extracellular interactions.
Eliminating the lack of drug targets in the development of marketed drugs.
This study introduces a cutting-edge method utilizing a proximity biotinylation enzyme, AirID, fused with an antigen-binding fragment (FabID) to explore extracellular protein-protein interactions (exPPIs) of the epidermal growth factor receptor (EGFR). By enabling precise biotinylation of proteins near EGFR on cell membranes, and subsequent analysis with LC-MS/MS, this approach uncovers both known and novel EGFR interactors. It highlights how these interactions dynamically change with EGF ligand and gefitinib treatment, offering new insights for receptor biology and drug development.
1. Background
Many proteins within the body form complexes with other proteins, determining the fate of cells. Therefore, the analysis of protein-protein interactions (PPI) is a crucial process for understanding the biological function of target proteins. Membrane proteins, which account for over 30% of human genes, play a vital role in cell functions. Many membrane proteins are known to form complexes to exert their functions, making the elucidation of membrane protein PPIs essential for understanding protein function. However, the development of technologies to analyze PPIs, especially extracellular protein–protein interactions (exPPIs) of living cells, has been lagging. Recently, proximity labeling methods that label proteins in close proximity for large-scale PPI analysis have gained attention. However, proximity protein labeling methods targeting the extracellular region of membrane proteins have primarily involved PPI analysis using molecules with cellular toxicity, prompting a search for systems targeting living cells.
2. Research Findings
The Proteo-Science Center at Ehime University has independently developed the proximity-dependent biotin labeling enzyme AirID (Kido, et al., eLife 2020), which biotinylates lysine residues of proteins in close proximity. There have been several studies using proximity-dependent biotin labeling enzymes for exPPI analysis, but they involved genetically modified proteins with significantly different shapes from their original structures, making it unclear to what extent the analysis results reflect the original interactions. To accurately understand interactions occurring on the cell membrane of living cells, it was necessary to develop a technology that could directly target proteins expressed by the cells for exPPI analysis. Therefore, our research group initiated this study on the premise that it would be possible to analyze exPPI by creating a molecule (FabID) that fuses AirID to the antigen recognition site of an antibody recognizing the extracellular domain of membrane proteins.
The membrane protein analyzed for exPPI was the epidermal growth factor receptor EGFR, a protein known as a cancer gene located on the cell membrane. When FabID and biotin were added to epithelial-like carcinoma-derived cells (A431 cells) (hereinafter referred to as epithelial cancer cells) expressing EGFR, it was confirmed at the cell culture level that EGFR on the cell membrane could be biotinylated using FabID.
By combining biotin labeling with FabID and mass spectrometry developed at Tokushima University for analyzing biotinylated proteins, we successfully identified many novel EGFR interacting proteins. The identified proteins have the potential to be new drug targets.
EGFR transmits signals to cells by binding to a ligand known as EGF. It is known that when EGF binds to EGFR, various proteins bind to the intracellular domain of EGFR, forming a protein complex. Currently, EGFR tyrosine kinase inhibitors, widely used as cancer treatments, are thought to exert their therapeutic effects by binding to EGFR and inhibiting the formation of protein complexes mediated by the EGFR intracellular domain. However, the ligand-dependent and drug-dependent exPPI changes of EGFR when EGFR binds to EGF (ligand) or when EGFR tyrosine kinase inhibitors (drugs) act have not been observed at all. Therefore, we observed ligand-dependent and drug-dependent exPPI changes of EGFR occurring in cultured cells using FabID. As a result, we are the first in the world to find that exPPI changes dynamically when ligands and drugs bind to EGFR.
3. Ripple Effect
Membrane proteins are used by almost all organisms to transmit information inside and outside cells. Therefore, analyzing the exPPI of membrane proteins is expected to directly contribute to the advancement of receptor biology. Moreover, since a majority of commercial drugs function by targeting membrane proteins, membrane proteins are known as important drug targets. However, finding new membrane protein drug targets is challenging, and it has been a significant issue in the pharmaceutical industry. The FabID technology developed in this study not only enables exPPI analysis using living cells but can also be used to identify novel drug targets. Furthermore, FabID has been found to capture ligand-dependent and drug-dependent exPPI changes that have not been analyzed by conventional methods. In the future, the identification of new drug targets through exPPI analysis using FabID and detailed analysis of membrane protein exPPI changes when commercial drugs bind are expected to contribute significantly to the development of commercial drugs.
4. Research Organization and Support
This research was conducted as a joint research project by the Proteo-Science Center at Ehime University, Institute of Advanced Medical Sciences at Tokushima University, the School of Life Sciences at Tokyo Pharmaceutical University, Department of Molecular Pharmacology of Tohoku University Graduate School of Medicine, and the Department of Bioscience of Nagahama Institute of BioScience and Technology.
Reference URL: https://www.nature.com/articles/s41467-023-43931-7
Bibliographic Information
Proximity extracellular protein-protein interaction analysis of EGFR using AirID-conjugated fragment of antigen binding, Kohdai Yamada, Ryouhei Shioya, Kohei Nishino, Hirotake Furihata, Atsushi Hijikata, Mika K. Kaneko, Yukinari Kato, Tsuyoshi Shirai, Hidetaka Kosako & Tatsuya Sawasaki, Nat Commun, 14(1):8301. doi: 10.1038/s41467-023-43931-7. 2023 Dec 14;
Fundings
- Basis for Supporting Innovative Drug Discovery and Life Science Research(JP21am0101077, 22ama121010j0001, 23ama121010j0002, JP22ama121008)
- JSPS KAKENHI(19H03218, JP21K19230)
- Takeda Science Foundation
Media
-
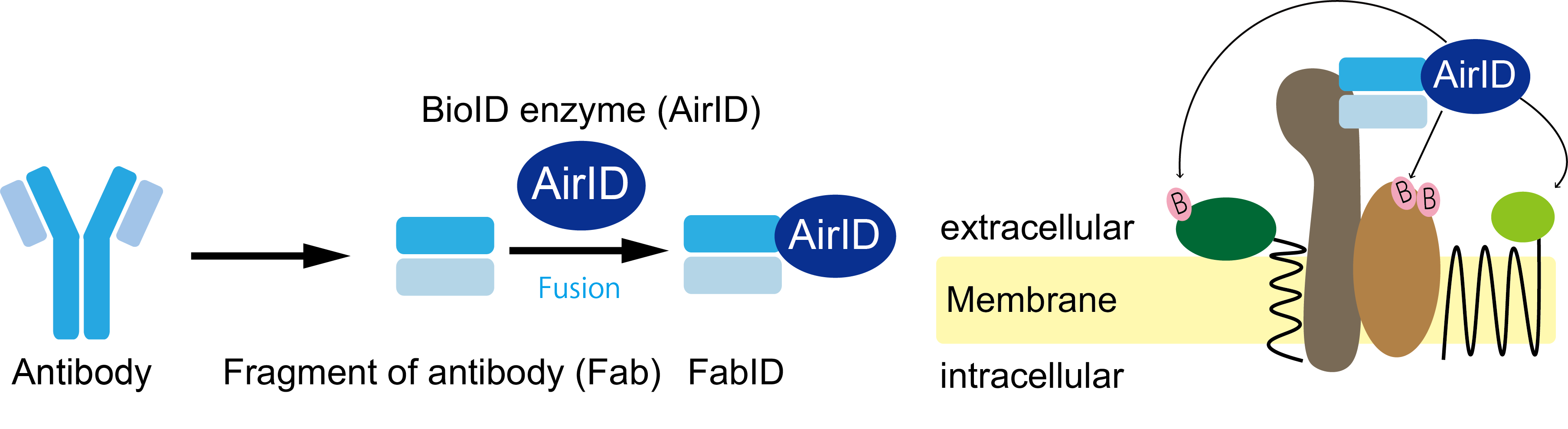
Overview of FabID technology developed in this study.
(left). Schematic diagram of AirID fused on the gene to the antigen-binding site of an antibody. FabID is a genetic fusion of AirID to the antigen binding site (right). Schematic diagram of membrane protein exPPI analysis using FabID. The molecule marked B in pink represents biotin, and proteins in close proximity to AirID are labelled biotinylated.
credit : Kohdai Yamada, Sawasaki Tatsuya
Usage Restriction : Please get copyright permission -
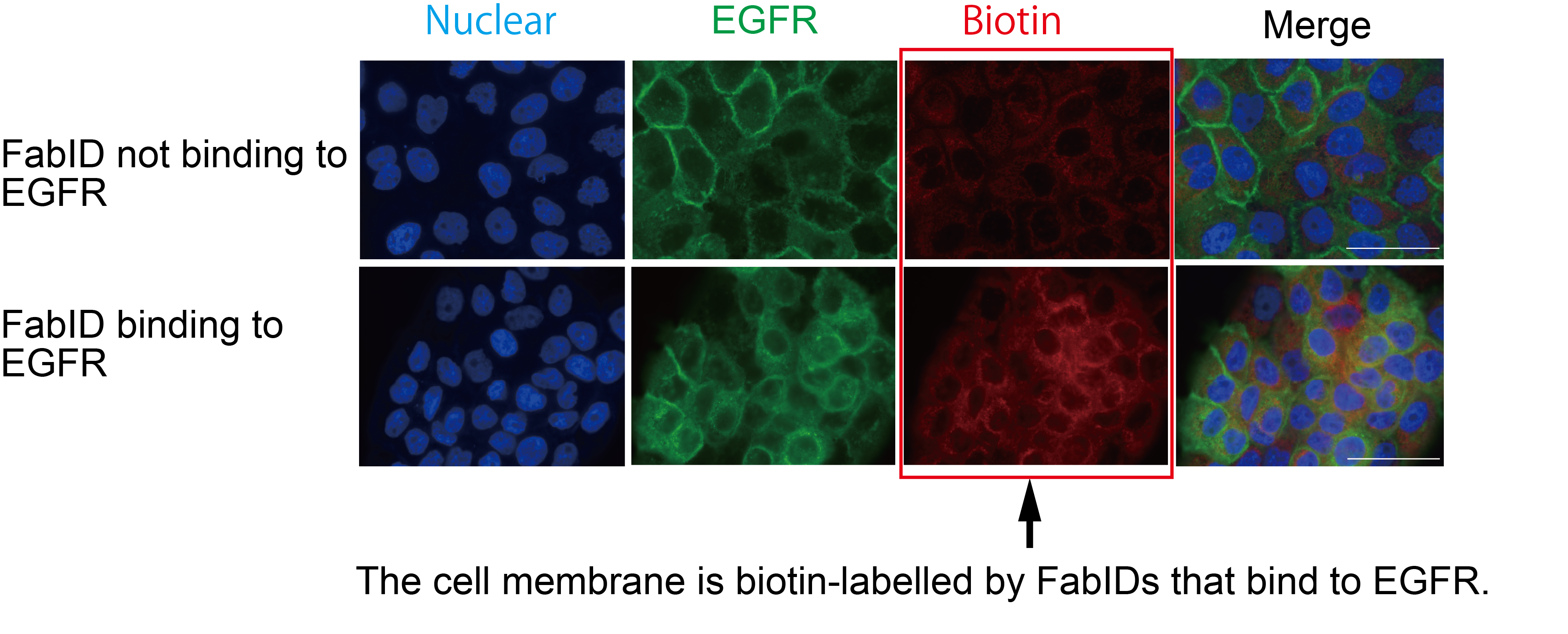
Immunostaining picture of FabID binding to EGFR on cultured cells, labelled on the plasma membrane.
Green represents the subcellular localization of EGFR and red represents biotinylated labelled proteins. Higher red color in the lower row indicates that the protein is more biotinylated. Cells were epithelial cancer cells (A431 cells).
credit : Kohdai Yamada, Sawasaki Tatsuya
Usage Restriction : Please get copyright permission -
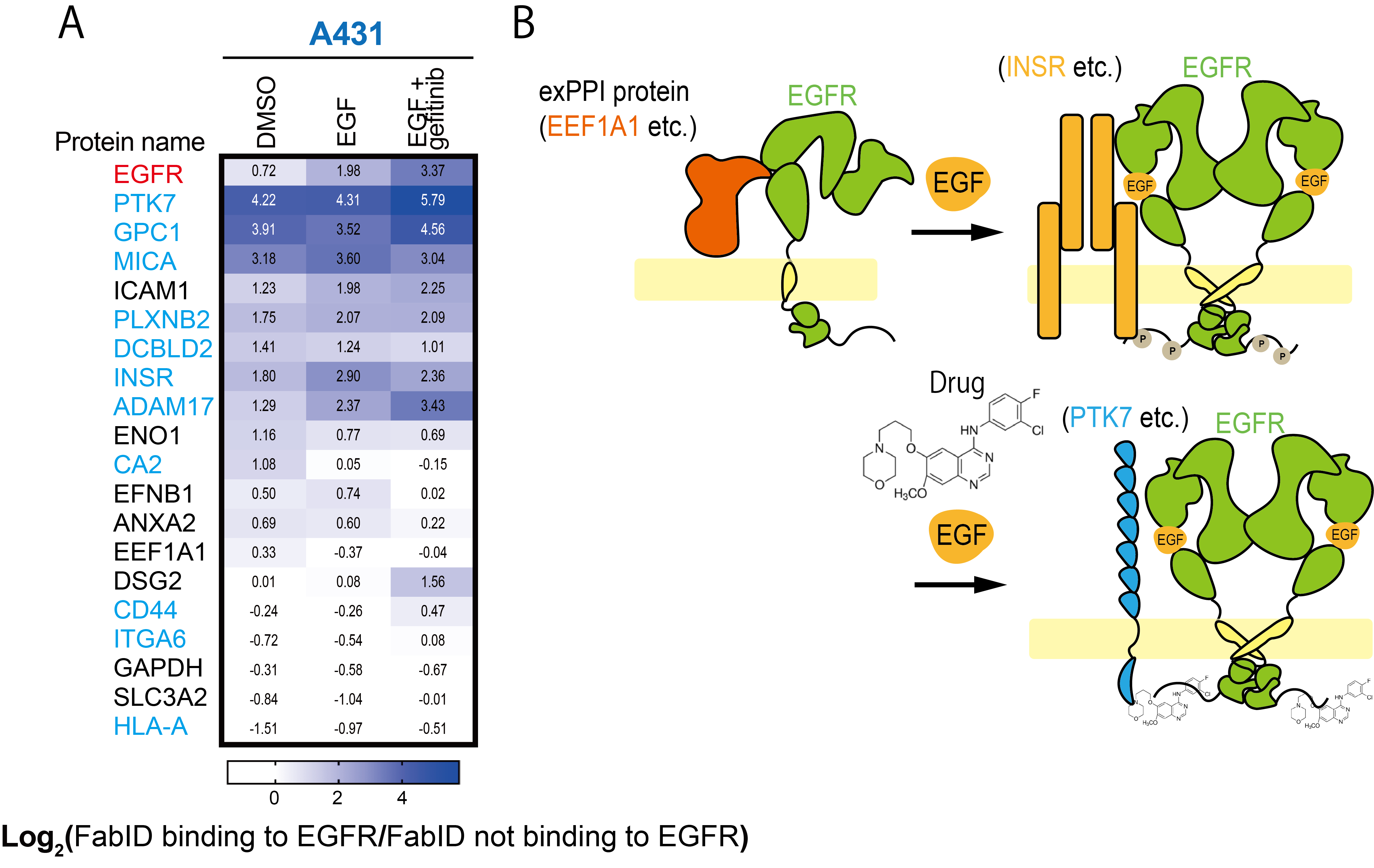
Changes in proximity labelled proteins due to binding of ligands and inhibitors to the EGFR.
A) Heatmap diagram showing biotinylation changes in DMSO (without ligand and drug addition), EGF (ligand) and EGF+Gefitinib (ligand + drug) treated zones. Cells were epithelial cancer cells (A431 cells). Black letters in proteins are known EGFR-interacting proteins, blue letters are EGFR-interacting proteins newly identified in the present study.
B) Diagram depicting exPPI changes in EGFR. In epithelial cancer cells, EGFR and EEF1A1 are in close proximity when EGF is not bound; EGF binding increases the degree of proximity between INSR and EGFR and drug binding there increases the degree of proximity between PTK7 and EGFR.credit : Kohdai Yamada, Sawasaki Tatsuya
Usage Restriction : Please get copyright permission
Contact Person
Name : Sawasaki Tatsuya
Phone : +81-89-927-8530
E-mail : sawasaki@ehime-u.ac.jp
Affiliation : Division of Cell-Free Life Science, Proteo-Science Center

